import rastereasy
Dealing with shapefiles, ROI, …
1) Read shapefile
Two main functions : rastereasy.shpfiles.shp2raster and rastereasy.shpfiles.shp2geoim
help(rastereasy.shpfiles.shp2raster)
help(rastereasy.shpfiles.shp2geoim)
Help on function shp2raster in module rastereasy:
shp2raster(shapefile_path, dest_name, attribute='code', resolution=10, nodata=0)
Convert a shapefile to a GeoTIFF raster file.
Parameters
----------
shapefile_path : str
Path to the input shapefile.
dest_name : str
Path to save the output raster file.
attribute : str, optional
Attribute field in the shapefile to assign values to each pixel.
Default is 'code'.
resolution : float, optional
Spatial resolution of the output raster in meters/degrees.
Default is 10.
nodata : int or float, optional
Value to assign to areas outside the shapes.
Default is 0.
Notes
-----
- The `shapefile_path` should be the full path to a shapefile (.shp) on the disk.
- To get the attributes of a shapefile, see :meth:`shpfiles.get_shapefile_attributes`
- The output raster will be written in GeoTIFF format to the path specified by `dest_name`.
Examples
--------
>>> shpfiles.shp2raster("landcover.shp", "landcover.tif", attribute='landtype', resolution=5)
Help on function shp2geoim in module rastereasy:
shp2geoim(shapefile_path, attribute='code', resolution=10, nodata=0)
Convert a shapefile to a Geoimage object.
Parameters
----------
shapefile_path : str
Path to the input shapefile.
attribute : str, optional
Attribute field in the shapefile to assign values to each pixel.
Default is 'code'.
resolution : float, optional
Spatial resolution of the output raster in meters/degrees.
Default is 10.
nodata : int or float, optional
Value to assign to areas outside the shapes.
Default is 0.
Returns
-------
Geoimage
A Geoimage object containing the rasterized data.
Notes
-----
- The `shapefile_path` should be the full path to a shapefile (.shp) on the disk.
- The `attribute` field will be assigned to each pixel in the rasterized Geoimage.
- To get the attributes of a shapefile, see :meth:`shpfiles.get_shapefile_attributes`
- The `resolution` sets the size of each pixel in the output image.
Examples
--------
>>> geo_img = shpfiles.shp2geoim("landcover.shp", attribute='landtype', resolution=5)
2) Convert shapefile to tif
shapefile_path = './data/demo/data_shp/Roi_G5.shp'
rastereasy.shpfiles.shp2raster(shapefile_path,'./data/results/shp2raster/raster5m.tif',resolution=5)
rastereasy.shpfiles.shp2raster(shapefile_path,'./data/results/shp2raster/raster10m.tif',resolution=10)
3) Import as Geoimage
raster10m=rastereasy.open('./data/results/shp2raster/raster10m.tif')
raster5m=rastereasy.open('./data/results/shp2raster/raster5m.tif')
raster5m.info()
raster10m.info()
print(raster10m.unique())
print(raster5m.unique())
raster10m.visu()
raster5m.visu()
- Size of the image:
- Rows (height): 3649
- Cols (width): 5097
- Bands: 1
- Spatial resolution: 5.000057261870794 meters / degree (depending on projection system)
- Central point latitude - longitude coordinates: (47.60953547, -2.57061505)
- Driver: GTiff
- Data type: int32
- Projection system: EPSG:32630
- Nodata: 0.0
- Given names for spectral bands:
{'1': 1}
- Size of the image:
- Rows (height): 1824
- Cols (width): 2548
- Bands: 1
- Spatial resolution: 10.002076869605746 meters / degree (depending on projection system)
- Central point latitude - longitude coordinates: (47.60951283, -2.57058196)
- Driver: GTiff
- Data type: int32
- Projection system: EPSG:32630
- Nodata: 0.0
- Given names for spectral bands:
{'1': 1}
[0 1 2 3 4 5 6 7]
[0 1 2 3 4 5 6 7]
4) Convert shapefile to geoimage
georaster5=rastereasy.shpfiles.shp2geoim(shapefile_path,resolution=5)
georaster10=rastereasy.shpfiles.shp2geoim(shapefile_path,resolution=10)
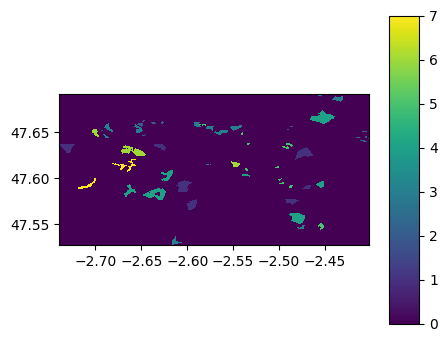
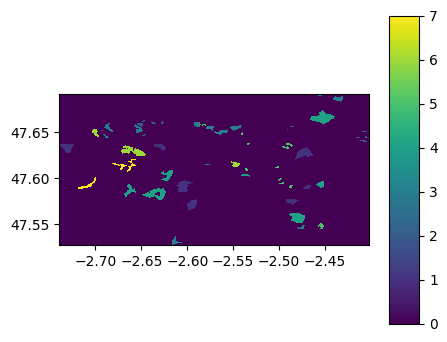
georaster5.visu(colorbar=True)
georaster10.visu(colorbar=True)
georaster5.save('./data/results/shp2raster/raster5m_fromgeoraster.tif')
georaster10.save('./data/results/shp2raster/raster10m_fromgeoraster.tif')
G5_B2=rastereasy.open('./data/demo/tostack/G5_B2.tif')
G5_B3=rastereasy.open('./data/demo/tostack/G5_B3.tif')
G5_B4=rastereasy.open('./data/demo/tostack/G5_B4.tif')
G5_B8=rastereasy.open('./data/demo/tostack/G5_B8.tif')
ROI_g5=rastereasy.open('./data/demo/data_shp/ROI_g5.tif')
ROI_g5.visu()
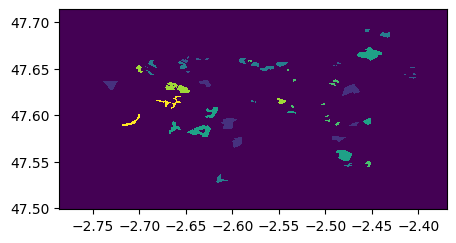
5) Select interesting areas with extract_from_shapefile
1) Without modifying the image
Read a stack (for the example. im can be an image)
im=rastereasy.open('./data/demo/tostack/G5_B2.tif')
im_stack = im.stack(rastereasy.open('./data/demo/tostack/G5_B3.tif'),reformat_names=True)
im_stack = im_stack.stack(rastereasy.open('./data/demo/tostack/G5_B4.tif'),reformat_names=True)
im_stack = im_stack.stack(rastereasy.open('./data/demo/tostack/G5_B8.tif'),reformat_names=True)
im_stack.info()
- Size of the image:
- Rows (height): 2370
- Cols (width): 3144
- Bands: 4
- Spatial resolution: 10.0 meters / degree (depending on projection system)
- Central point latitude - longitude coordinates: (47.60615500, -2.57780166)
- Driver: GTiff
- Data type: uint16
- Projection system: EPSG:32630
- Given names for spectral bands:
{'1': 1, '2': 2, '3': 3, '4': 4}
keep_size=True
value=3
ime=im_stack.extract_from_shapefile(shapefile_path,value,keep_size=keep_size)
ime.info()
- Size of the image:
- Rows (height): 2370
- Cols (width): 3144
- Bands: 4
- Spatial resolution: 10.0 meters / degree (depending on projection system)
- Central point latitude - longitude coordinates: (47.60615500, -2.57780166)
- Driver: GTiff
- Data type: uint16
- Projection system: EPSG:32630
- Nodata: 0
- Given names for spectral bands:
{'1': 1, '2': 2, '3': 3, '4': 4}
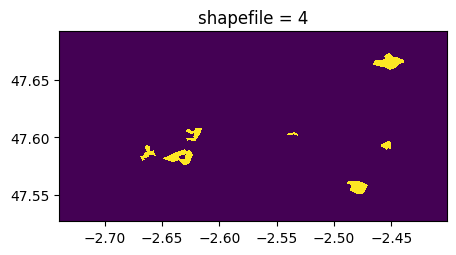
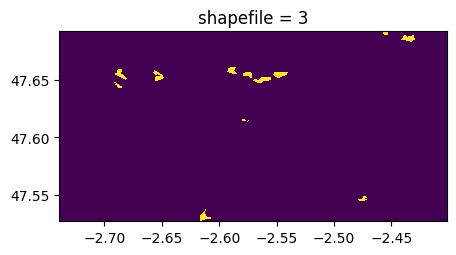
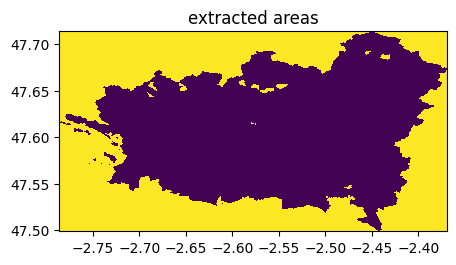
(rastereasy.shpfiles.shp2geoim(shapefile_path,resolution=im.resolution)==value).visu(title='shapefile = %d'%value)
if keep_size is True:
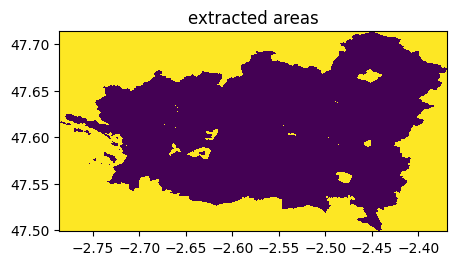
((ime-im)==0).visu(1,title='extracted areas')
else:
ime,imc = rastereasy.extract_common_areas(ime,im,resolution=None)
((ime-imc)==0).visu(1,title='extracted areas')
2) With modifying the image (option inplace=True)
keep_size=False
value=4
#Read a stack
im=rastereasy.open('./data/demo/tostack/G5_B2.tif')
im.stack(rastereasy.open('./data/demo/tostack/G5_B3.tif'),reformat_names=True, inplace=True)
im.stack(rastereasy.open('./data/demo/tostack/G5_B4.tif'),reformat_names=True, inplace=True)
im.stack(rastereasy.open('./data/demo/tostack/G5_B8.tif'),reformat_names=True, inplace=True)
imoriginal=im.copy()
imoriginal.info()
im.extract_from_shapefile(shapefile_path,value,keep_size=keep_size,inplace=True)
im.save('./data/results/shp2raster/extract4.tif')
im.info()
(rastereasy.shpfiles.shp2geoim(shapefile_path,resolution=im.resolution)==value).visu(title='shapefile = %d'%value)
if keep_size is True:
((imoriginal-im)==0).visu(1,title='extracted areas')
else:
ime,imc = rastereasy.extract_common_areas(imoriginal,im,resolution=None)
((ime-imc)==0).visu(1,title='extracted areas')
- Size of the image:
- Rows (height): 2370
- Cols (width): 3144
- Bands: 4
- Spatial resolution: 10.0 meters / degree (depending on projection system)
- Central point latitude - longitude coordinates: (47.60615500, -2.57780166)
- Driver: GTiff
- Data type: uint16
- Projection system: EPSG:32630
- Given names for spectral bands:
{'1': 1, '2': 2, '3': 3, '4': 4}
- Size of the image:
- Rows (height): 1824
- Cols (width): 2548
- Bands: 4
- Spatial resolution: 10.0 meters / degree (depending on projection system)
- Central point latitude - longitude coordinates: (47.60943270, -2.57056349)
- Driver: GTiff
- Data type: uint16
- Projection system: EPSG:32630
- Nodata: 0
- Given names for spectral bands:
{'1': 1, '2': 2, '3': 3, '4': 4}
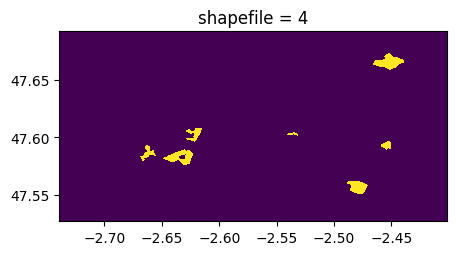
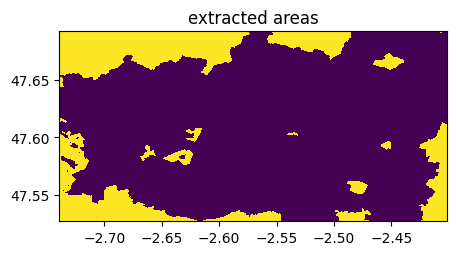
3) By having charged the shapefile
1) Without modifying the image
#Read a stack
shapefile_path = './data/demo/data_shp/Roi_G5.shp'
shp=rastereasy.shpfiles.shp2geoim(shapefile_path,resolution=10)
im=rastereasy.open('./data/demo/tostack/G5_B2.tif')
im_stack = im.stack(rastereasy.open('./data/demo/tostack/G5_B3.tif'),reformat_names=True)
im_stack = im_stack.stack(rastereasy.open('./data/demo/tostack/G5_B4.tif'),reformat_names=True)
im_stack = im_stack.stack(rastereasy.open('./data/demo/tostack/G5_B8.tif'),reformat_names=True)
im_stack.info()
- Size of the image:
- Rows (height): 2370
- Cols (width): 3144
- Bands: 4
- Spatial resolution: 10.0 meters / degree (depending on projection system)
- Central point latitude - longitude coordinates: (47.60615500, -2.57780166)
- Driver: GTiff
- Data type: uint16
- Projection system: EPSG:32630
- Given names for spectral bands:
{'1': 1, '2': 2, '3': 3, '4': 4}
keep_size=True
value=3
ime=im_stack.extract_from_shapeimage(shp,value,keep_size=keep_size)
ime.save('./data/results/shp2raster//extract3.tif')
ime.info()
- Size of the image:
- Rows (height): 2370
- Cols (width): 3144
- Bands: 4
- Spatial resolution: 10.0 meters / degree (depending on projection system)
- Central point latitude - longitude coordinates: (47.60615500, -2.57780166)
- Driver: GTiff
- Data type: uint16
- Projection system: EPSG:32630
- Nodata: 0
- Given names for spectral bands:
{'1': 1, '2': 2, '3': 3, '4': 4}
(rastereasy.shpfiles.shp2geoim(shapefile_path,resolution=im.resolution)==value).visu(title='shapefile = %d'%value)
if keep_size is True:
((ime-im)==0).visu(1,title='extracted areas')
else:
ime,imc = rastereasy.extract_common_areas(ime,im,resolution=None)
((ime-imc)==0).visu(1,title='extracted areas')


2) With modifying the image
shp=rastereasy.shpfiles.shp2geoim(shapefile_path,resolution=3)
keep_size=True
value=4
#Read a stack
im=rastereasy.open('./data/demo/tostack/G5_B2.tif')
im.stack(rastereasy.open('./data/demo/tostack/G5_B3.tif'),reformat_names=True, inplace=True)
im.stack(rastereasy.open('./data/demo/tostack/G5_B4.tif'),reformat_names=True, inplace=True)
im.stack(rastereasy.open('./data/demo/tostack/G5_B8.tif'),reformat_names=True, inplace=True)
imoriginal=im.copy()
imoriginal.info()
im.extract_from_shapeimage(shp,value,keep_size=keep_size,inplace=True)
im.save('./data/results/shp2raster//extract4.tif')
im.info()
(shp==value).visu(title='shapefile = %d'%value)
if keep_size is True:
((imoriginal-im)==0).visu(1,title='extracted areas')
else:
ime,imc = rastereasy.extract_common_areas(imoriginal,im,resolution=None)
((ime-imc)==0).visu(1,title='extracted areas')
- Size of the image:
- Rows (height): 2370
- Cols (width): 3144
- Bands: 4
- Spatial resolution: 10.0 meters / degree (depending on projection system)
- Central point latitude - longitude coordinates: (47.60615500, -2.57780166)
- Driver: GTiff
- Data type: uint16
- Projection system: EPSG:32630
- Given names for spectral bands:
{'1': 1, '2': 2, '3': 3, '4': 4}
- Size of the image:
- Rows (height): 2370
- Cols (width): 3144
- Bands: 4
- Spatial resolution: 10.0 meters / degree (depending on projection system)
- Central point latitude - longitude coordinates: (47.60615500, -2.57780166)
- Driver: GTiff
- Data type: uint16
- Projection system: EPSG:32630
- Nodata: 0
- Given names for spectral bands:
{'1': 1, '2': 2, '3': 3, '4': 4}