import rastereasy
name_im='./data/demo/sentinel.tif'
image=rastereasy.open(name_im)
Extract numpy arrays
1) Channel first format
im=image.numpy_channel_first()
print(im.shape)
(12, 1000, 1000)
### 2) Channel last format
im=image.numpy_channel_last()
print(im.shape)
(1000, 1000, 12)
Some channels in numpy array
a) If names are not given
bands=["1","4","3"]
im=image.numpy_channel_first(bands=bands)
print(im.shape)
bands=[1,4,3,8]
im=image.numpy_channel_last(bands=bands)
print(im.shape)
(3, 1000, 1000)
(1000, 1000, 4)
b) If names are given
# first : change the names of the image
names = {"NIR":8,"G":3,"CO" : 1,"SWIR2":11,"B": 2,"R":4,"RE1":5,"RE2":6,"RE3":7,"WA":9,"SWIR1":10,"SWIR3":12}
image.change_names(names)
image.info()
- Size of the image:
- Rows (height): 1000
- Cols (width): 1000
- Bands: 12
- Spatial resolution: 10.0 meters / degree (depending on projection system)
- Central point latitude - longitude coordinates: (7.04099599, 38.39058840)
- Driver: GTiff
- Data type: int16
- Projection system: EPSG:32637
- Nodata: -32768.0
- Given names for spectral bands:
{'CO': 1, 'B': 2, 'G': 3, 'R': 4, 'RE1': 5, 'RE2': 6, 'RE3': 7, 'NIR': 8, 'WA': 9, 'SWIR1': 10, 'SWIR2': 11, 'SWIR3': 12}
bands=["R","G","B"]
im=image.numpy_channel_first(bands=bands)
print(im.shape)
bands=["R","NIR","G","B"]
im=image.numpy_channel_last(bands=bands)
print(im.shape)
(3, 1000, 1000)
(1000, 1000, 4)
Entire image in table of size (row*col, bands)
help(image.numpy_table)
Help on method numpy_table in module rastereasy:
numpy_table(bands=None) method of rastereasy.Geoimage instance
Extract image data as a 2D table of shape (pixels, bands).
This method reshapes the image into a 2D table where each row represents a pixel
and each column represents a band. This format is useful for machine learning,
statistical analysis, or any operation that treats pixels as independent samples.
Parameters
----------
bands : str, list of str, or None, optional
The bands to include in the output:
- If None: Returns all bands
- If a string: Returns a single specified band
- If a list: Returns the specified bands in the given order
Default is None.
Returns
-------
numpy.ndarray
Image data as a 2D table with shape (rows*cols, bands)
Examples
--------
>>> # Convert the entire image to a table
>>> table = image.numpy_table()
>>> print(f"Table shape: {table.shape}")
>>>
>>> # Process specific bands as a table
>>> nir_red = image.numpy_table(bands=["NIR", "R"])
>>> print(f"Shape: {nir_red.shape}")
>>> ndvi = (nir_red[:, 0] - nir_red[:, 1]) / (nir_red[:, 0] + nir_red[:, 1])
>>> print(f"Mean NDVI: {ndvi.mean()}")
Notes
-----
This format is particularly useful for:
- Machine learning where each pixel is a sample and each band is a feature
- Clustering algorithms like K-means
- Statistical analysis across bands
- Vectorized operations on pixels
table=image.numpy_table()
print(table.shape)
(1000000, 12)
Recovering a numpy array (channel_first or channel_last) from a table
help(rastereasy.table2image)
Help on function table2image in module rastereasy:
table2image(table, size, channel_first=True)
Reshape a 2D table back into a 3D image.
Parameters
----------
table : numpy.ndarray
Input table with shape (rows*cols, bands) or (rows*cols,).
size : tuple
Size of the output image as (rows, cols).
channel_first : bool, optional
If True, output will have shape (bands, rows, cols).
If False, output will have shape (rows, cols, bands).
Default is True.
Returns
-------
numpy.ndarray
Reshaped 3D image.
Examples
--------
>>> image = table2image(table, (400, 600), channel_first=True)
# Test with channel first recovery
image_recovered=rastereasy.table2image(table,image.shape)
image_recovered.shape
(12, 1000, 1000)
# check the consistency
import numpy as np
print(np.sum(np.abs(image_recovered-image.numpy_channel_first())))
print(image_recovered.shape)
# Test with channel last recovery
image_recovered=rastereasy.table2image(table,image.shape,channel_first=False)
print(np.sum(np.abs(image_recovered-image.numpy_channel_last())))
print(image_recovered.shape)
0
(12, 1000, 1000)
0
(1000, 1000, 12)
Creating a Geoimage from a table
We can directly create a geoimage from the table. Dimensions should match with function image_from_table
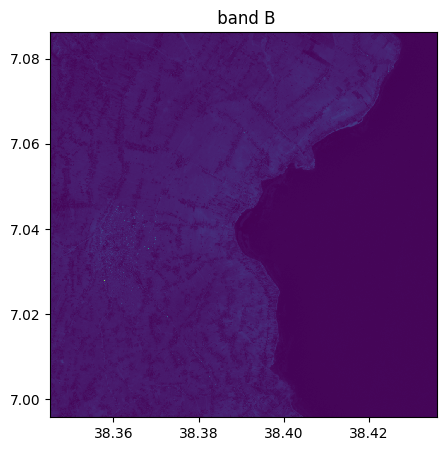
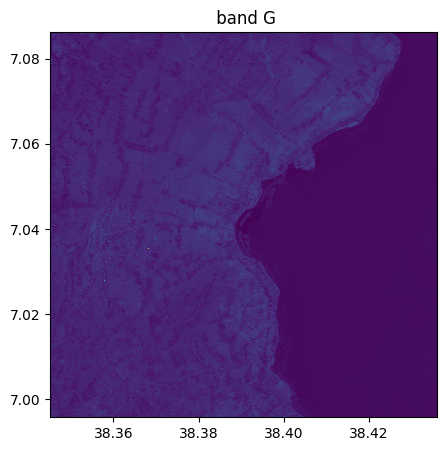
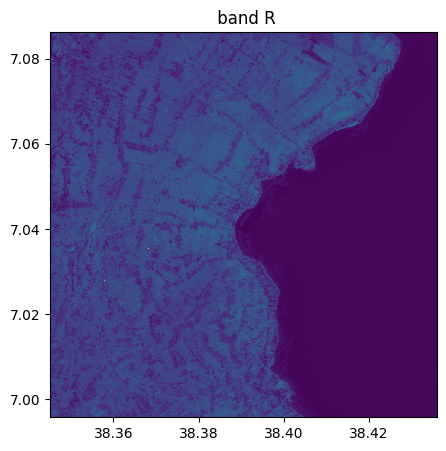
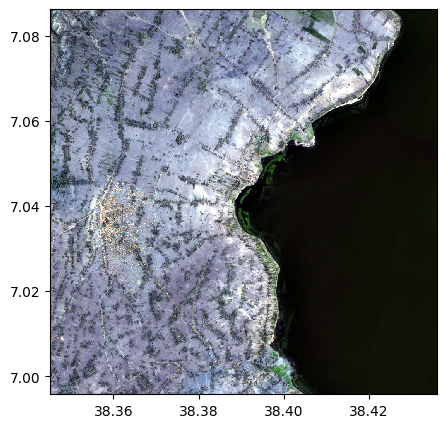
image_recovered=image.image_from_table(table[:,1:4],names={"R":3,"G":2,"B":1})
image_recovered.info()
image_recovered.visu()
image_recovered.colorcomp()
- Size of the image:
- Rows (height): 1000
- Cols (width): 1000
- Bands: 3
- Spatial resolution: 10.0 meters / degree (depending on projection system)
- Central point latitude - longitude coordinates: (7.04099599, 38.39058840)
- Driver: GTiff
- Data type: int16
- Projection system: EPSG:32637
- Nodata: -32768.0
- Given names for spectral bands:
{'B': 1, 'G': 2, 'R': 3}
Creating a Geoimage from a numpy array (channel_first or channel_last)
We can directly create a geoimage from a numpy array with function upload_image.
bands=["R","B","SWIR1"]
im_np=image.numpy_channel_first(bands=bands)
print(im_np.shape)
(3, 1000, 1000)
# We modify bands
im_np[2,:,:]=im_np[1,:,:]+im_np[0,:,:]
We put the new image with the modified 3 bands in a Geoimage
help(image.upload_image)
Help on method upload_image in module rastereasy:
upload_image(
image,
names=None,
dest_name=None,
channel_first=True,
inplace=False
) method of rastereasy.Geoimage instance
Update the image data with a new image array.
This method replaces the current image data with a new image array. The new image
must have compatible dimensions with the current image.
Parameters
----------
image : numpy.ndarray
The new image data to upload, with shape:
- (bands, rows, cols) if channel_first=True
- (rows, cols, bands) if channel_first=False
- (rows, cols) for a single band
names : dict, optional
Dictionary mapping band names to band indices. If None, bands will be named
sequentially ("1", "2", "3", ...).
Default is None.
dest_name : str, optional
Path to save the updated image. If None, the image is not saved.
Default is None.
channel_first : bool, optional
Whether the input image has channels in the first dimension (True) or the last
dimension (False).
Default is True.
inplace : bool, default False
If False, return a copy. Otherwise, upload image in place and return None.
Returns
-------
Geoimage
The updated image or None if `inplace=True`
Raises
------
ValueError
If the spatial dimensions of the new image don't match the original image
Examples
--------
Examples
--------
>>> # Create a new filtered image without modifying the original
>>> array = image.numpy_channel_first()
>>> filtered = apply_some_filter(array) # Apply some processing
>>> filtered_image = image.upload_image(filtered)
>>> filtered_image.visu()
>>> image.visu() # Original remains unchanged
>>>
>>> # Create a single-band image from NDVI calculation
>>> nir = image.numpy_channel_first(bands=["NIR"])
>>> red = image.numpy_channel_first(bands=["Red"])
>>> ndvi = (nir - red) / (nir + red)
>>> ndvi_image = image.upload_image(ndvi, names={"NDVI": 1},
>>> dest_name="ndvi.tif")
>>> # Update an image with processed data
>>> array = image.numpy_channel_first()
>>> filtered = some_filter_function(array) # Apply some processing
>>> image.upload_image(filtered, inplace=True)
>>> image.visu()
>>>
>>> # Upload an image in channel-last format
>>> import cv2
>>> bgr = cv2.imread('rgb.jpg') # OpenCV uses BGR order
>>> rgb = bgr[:, :, ::-1] # Convert BGR to RGB
>>> image.upload_image(rgb, channel_first=False, dest_name="from_jpeg.tif", inplace=True))
Notes
-----
The spatial dimensions (rows, cols)
must match the original image, but the number of bands can change.
image_modified=image.upload_image(im_np)
image_modified.info()
- Size of the image:
- Rows (height): 1000
- Cols (width): 1000
- Bands: 3
- Spatial resolution: 10.0 meters / degree (depending on projection system)
- Central point latitude - longitude coordinates: (7.04099599, 38.39058840)
- Driver: GTiff
- Data type: int16
- Projection system: EPSG:32637
- Nodata: -32768.0
- Given names for spectral bands:
{'1': 1, '2': 2, '3': 3}
image.info()
- Size of the image:
- Rows (height): 1000
- Cols (width): 1000
- Bands: 12
- Spatial resolution: 10.0 meters / degree (depending on projection system)
- Central point latitude - longitude coordinates: (7.04099599, 38.39058840)
- Driver: GTiff
- Data type: int16
- Projection system: EPSG:32637
- Nodata: -32768.0
- Given names for spectral bands:
{'CO': 1, 'B': 2, 'G': 3, 'R': 4, 'RE1': 5, 'RE2': 6, 'RE3': 7, 'NIR': 8, 'WA': 9, 'SWIR1': 10, 'SWIR2': 11, 'SWIR3': 12}